Training a transfer learning model#
This tutorial illustrates how to use a transfer learning approach to train a model on the GC mEPSP dataset. The miniML base model, trained on GC mEPSCs, is used as a starting point. We freeze the convolutional layers of the base model and train the remaining layers on the GC mEPSP dataset. This transfer learning approach greatly reduces the training time and the required size of the training dataset.
Preparation#
We begin by downloading the training dataset for training a miniML mEPSP detection model from Zenodo. The dataset contains labeled data segments of mEPSPs and non-events.
import requests
file_url = 'https://zenodo.org/records/14507343/files/2_GC_mepsp_train.h5'
response = requests.get(file_url)
if response.status_code == 200:
with open('../_data/GC_mEPSP_training_data.h5', 'wb') as file:
file.write(response.content)
print('Downloaded GC_mEPSP_training_data.h5')
Downloaded GC_mEPSP_training_data.h5
Now we set up our Python environment:
import tensorflow as tf
from sklearn.preprocessing import minmax_scale
from sklearn.model_selection import train_test_split
from sklearn.metrics import auc, confusion_matrix, ConfusionMatrixDisplay, roc_curve
from tensorflow.keras.layers import (Input, BatchNormalization, AveragePooling1D, Conv1D,
Bidirectional, LSTM, Dense, Dropout, LeakyReLU)
from tensorflow.keras.optimizers.legacy import Adam
from scipy.signal import resample
import h5py
import numpy as np
import time
import matplotlib.pyplot as plt
import warnings
print('Using TF version', tf.__version__)
print('GPUs available:', len(tf.config.list_physical_devices('GPU')))
Using TF version 2.15.0
GPUs available: 1
Set training parameters#
We set the training parameters in a dictionary called settings.
settings = {
'training_size': 0.8,
'testing_size': None,
'learn_rate': 2e-8,
'epsilon': 1e-8,
'patience': 12,
'epochs': 100,
'batch_size': 64,
'dropout': 0.5,
'training_data': '../_data/GC_mEPSP_training_data.h5',
'base_model': '../../models/GC_lstm_model.h5'
}
Prepare training data#
Now we can start by loading the training dataset. The data is stored in a HDF5 file.
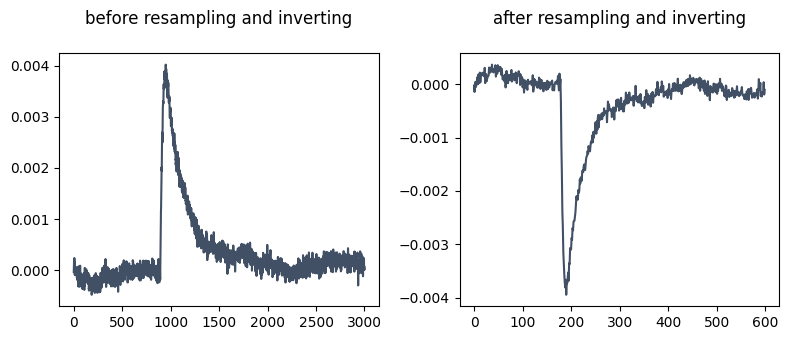
The data is loaded into the x and y variables. The data is resampled to 600 datapoints and the events are inverted. Because the base model was trained to detect downward deflections, inverting the data helps when training on data having a different direction. When using the trained model, remember to set the event_direction parameter to ‘positive’ when generating the EventDetection object.
with h5py.File(f'{settings["training_data"]}', 'r') as f:
x = f['events'][:]
y = f['scores'][:]
fig, axs = plt.subplots(1, 2, figsize=(8,3.5))
axs[0].plot(x[505], c='#425066')
axs[0].set_title('before resampling and inverting\n')
# resample and invert the data
x = resample(x, 600, axis=1)
x *= -1
axs[1].plot(x[505], c='#425066')
axs[1].set_title('after resampling and inverting\n')
plt.tight_layout()
plt.show()
Next, we proceed as for full model training by min-max scaling and splitting the data into training and testing sets.
scaled_data = minmax_scale(x, feature_range=(0,1), axis=1)
scaled_data = np.expand_dims(scaled_data, axis=2)
merged_y = np.expand_dims(y, axis=1)
print(f'loaded events with shape {scaled_data.shape}')
print(f'loaded scores with shape {merged_y.shape}')
print(f'ratio of pos/neg scores: {merged_y.sum()/(merged_y.shape[0]-merged_y.sum()):.2f}')
if merged_y.sum()/(merged_y.shape[0]-merged_y.sum()) > 1.05:
warnings.warn("unbalanced dataset: ratio of positive and negative scores >1.05")
elif merged_y.sum()/(merged_y.shape[0]-merged_y.sum()) < 0.95:
warnings.warn("unbalanced dataset: ratio of positive and negative scores < 0.95")
x_train, x_test, y_train, y_test = train_test_split(scaled_data, merged_y, train_size=settings['training_size'], random_state=1234)
loaded events with shape (4004, 600, 1)
loaded scores with shape (4004, 1)
ratio of pos/neg scores: 1.01
Prepare the model#
def build_model(x_train, dropout_rate):
model = tf.keras.models.Sequential()
model.add(Input(shape=(x_train.shape[1:])))
model.add(Conv1D(filters=32, kernel_size=9, padding='same'))
model.add(BatchNormalization())
model.add(LeakyReLU())
model.add(AveragePooling1D(pool_size=3, strides=3))
model.add(Conv1D(filters=48, kernel_size=7, padding='same'))
model.add(BatchNormalization())
model.add(LeakyReLU())
model.add(AveragePooling1D(pool_size=2, strides=2))
model.add(Conv1D(filters=64, kernel_size=5, padding='same'))
model.add(BatchNormalization())
model.add(LeakyReLU())
model.add(AveragePooling1D(pool_size=2, strides=2))
model.add(Conv1D(filters=80, kernel_size=3, padding='same'))
model.add(BatchNormalization())
model.add(LeakyReLU())
model.add(Bidirectional(LSTM(96, dropout=dropout_rate), merge_mode='sum'))
model.add(Dense(128, activation=LeakyReLU()))
model.add(Dropout(dropout_rate))
model.add(Dense(1, activation='sigmoid'))
return model
Next, we save the model weights from the pre-trained miniML model (the GC mEPSC base model). We then create a new model instance and load the saved weights.
old_model = tf.keras.models.load_model(settings['base_model'], compile=True)
old_model.save_weights('../_data/gc_weights')
new_model = build_model(x_train, settings['dropout'])
new_model.load_weights('../_data/gc_weights')
2025-03-21 17:31:19.177930: I metal_plugin/src/device/metal_device.cc:1154] Metal device set to: Apple M1 Pro
2025-03-21 17:31:19.177955: I metal_plugin/src/device/metal_device.cc:296] systemMemory: 16.00 GB
2025-03-21 17:31:19.177961: I metal_plugin/src/device/metal_device.cc:313] maxCacheSize: 5.33 GB
2025-03-21 17:31:19.177991: I tensorflow/core/common_runtime/pluggable_device/pluggable_device_factory.cc:306] Could not identify NUMA node of platform GPU ID 0, defaulting to 0. Your kernel may not have been built with NUMA support.
2025-03-21 17:31:19.178003: I tensorflow/core/common_runtime/pluggable_device/pluggable_device_factory.cc:272] Created TensorFlow device (/job:localhost/replica:0/task:0/device:GPU:0 with 0 MB memory) -> physical PluggableDevice (device: 0, name: METAL, pci bus id: <undefined>)
<tensorflow.python.checkpoint.checkpoint.CheckpointLoadStatus at 0x34cd23670>
For transfer learning, we have to freeze the convolutional layers of our model to prevent them from being trained. Only the remaining layers remain trainable.
for ind, layer in enumerate(new_model.layers):
if layer.name == 'BatchNormalization':
new_model.layers[ind].trainable = False
else:
if ind < len(new_model.layers) - 4:
new_model.layers[ind].trainable = False
else:
new_model.layers[ind].trainable = True
new_model.compile(optimizer=Adam(learning_rate=settings['learn_rate'], epsilon=settings['epsilon'], amsgrad=True),
loss=tf.keras.losses.BinaryCrossentropy(),
metrics=['Accuracy'])
new_model.summary(show_trainable=True)
Model: "sequential"
____________________________________________________________________________
Layer (type) Output Shape Param # Trainable
============================================================================
conv1d (Conv1D) (None, 600, 32) 320 N
batch_normalization (Batch (None, 600, 32) 128 N
Normalization)
leaky_re_lu (LeakyReLU) (None, 600, 32) 0 N
average_pooling1d (Average (None, 200, 32) 0 N
Pooling1D)
conv1d_1 (Conv1D) (None, 200, 48) 10800 N
batch_normalization_1 (Bat (None, 200, 48) 192 N
chNormalization)
leaky_re_lu_1 (LeakyReLU) (None, 200, 48) 0 N
average_pooling1d_1 (Avera (None, 100, 48) 0 N
gePooling1D)
conv1d_2 (Conv1D) (None, 100, 64) 15424 N
batch_normalization_2 (Bat (None, 100, 64) 256 N
chNormalization)
leaky_re_lu_2 (LeakyReLU) (None, 100, 64) 0 N
average_pooling1d_2 (Avera (None, 50, 64) 0 N
gePooling1D)
conv1d_3 (Conv1D) (None, 50, 80) 15440 N
batch_normalization_3 (Bat (None, 50, 80) 320 N
chNormalization)
leaky_re_lu_3 (LeakyReLU) (None, 50, 80) 0 N
bidirectional (Bidirection (None, 96) 135936 Y
al)
dense (Dense) (None, 128) 12416 Y
dropout (Dropout) (None, 128) 0 Y
dense_1 (Dense) (None, 1) 129 Y
============================================================================
Total params: 191361 (747.50 KB)
Trainable params: 148481 (580.00 KB)
Non-trainable params: 42880 (167.50 KB)
____________________________________________________________________________
Training#
Now we can train our model using the mEPSP dataset. Only the previously selected layers are trained. Because we have initialized the model weights from the GC mEPSC base model, the training is also faster than for full model training.
early_stopping_callback = tf.keras.callbacks.EarlyStopping(
monitor='val_loss',
patience=settings['patience'],
restore_best_weights=True)
start = time.time()
history = new_model.fit(x_train, y_train,
verbose=2,
epochs=settings['epochs'],
batch_size=settings['batch_size'],
validation_data=(x_test, y_test),
shuffle=True,
callbacks=[early_stopping_callback])
print('')
print('----')
print(f'train shape: {x_train.shape}')
print(f'score on val: {new_model.evaluate(x_test, y_test, verbose=0)[1]}')
print(f'score on train: {new_model.evaluate(x_train, y_train, verbose=0)[1]}')
print('----')
print(f'training time (s): {time.time()-start:.4f}')
Epoch 1/100
2025-03-21 17:31:21.050725: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:117] Plugin optimizer for device_type GPU is enabled.
51/51 - 4s - loss: 0.1969 - Accuracy: 0.9238 - val_loss: 0.2696 - val_Accuracy: 0.9238 - 4s/epoch - 87ms/step
Epoch 2/100
51/51 - 1s - loss: 0.1658 - Accuracy: 0.9341 - val_loss: 0.2314 - val_Accuracy: 0.9326 - 1s/epoch - 25ms/step
Epoch 3/100
51/51 - 1s - loss: 0.1560 - Accuracy: 0.9360 - val_loss: 0.2118 - val_Accuracy: 0.9351 - 1s/epoch - 25ms/step
Epoch 4/100
51/51 - 1s - loss: 0.1491 - Accuracy: 0.9404 - val_loss: 0.2232 - val_Accuracy: 0.9326 - 1s/epoch - 25ms/step
Epoch 5/100
51/51 - 1s - loss: 0.1416 - Accuracy: 0.9404 - val_loss: 0.2089 - val_Accuracy: 0.9351 - 1s/epoch - 26ms/step
Epoch 6/100
51/51 - 1s - loss: 0.1444 - Accuracy: 0.9410 - val_loss: 0.1903 - val_Accuracy: 0.9338 - 1s/epoch - 26ms/step
Epoch 7/100
51/51 - 1s - loss: 0.1328 - Accuracy: 0.9429 - val_loss: 0.1786 - val_Accuracy: 0.9338 - 1s/epoch - 26ms/step
Epoch 8/100
51/51 - 1s - loss: 0.1327 - Accuracy: 0.9482 - val_loss: 0.1753 - val_Accuracy: 0.9351 - 1s/epoch - 25ms/step
Epoch 9/100
51/51 - 1s - loss: 0.1253 - Accuracy: 0.9472 - val_loss: 0.1587 - val_Accuracy: 0.9326 - 1s/epoch - 26ms/step
Epoch 10/100
51/51 - 1s - loss: 0.1215 - Accuracy: 0.9500 - val_loss: 0.1751 - val_Accuracy: 0.9338 - 1s/epoch - 26ms/step
Epoch 11/100
51/51 - 1s - loss: 0.1211 - Accuracy: 0.9507 - val_loss: 0.1671 - val_Accuracy: 0.9338 - 1s/epoch - 26ms/step
Epoch 12/100
51/51 - 1s - loss: 0.1166 - Accuracy: 0.9479 - val_loss: 0.1458 - val_Accuracy: 0.9388 - 1s/epoch - 26ms/step
Epoch 13/100
51/51 - 1s - loss: 0.1212 - Accuracy: 0.9510 - val_loss: 0.1487 - val_Accuracy: 0.9388 - 1s/epoch - 26ms/step
Epoch 14/100
51/51 - 1s - loss: 0.1222 - Accuracy: 0.9491 - val_loss: 0.1470 - val_Accuracy: 0.9413 - 1s/epoch - 26ms/step
Epoch 15/100
51/51 - 1s - loss: 0.1219 - Accuracy: 0.9472 - val_loss: 0.1427 - val_Accuracy: 0.9413 - 1s/epoch - 26ms/step
Epoch 16/100
51/51 - 1s - loss: 0.1167 - Accuracy: 0.9504 - val_loss: 0.1485 - val_Accuracy: 0.9413 - 1s/epoch - 26ms/step
Epoch 17/100
51/51 - 1s - loss: 0.1109 - Accuracy: 0.9525 - val_loss: 0.1544 - val_Accuracy: 0.9413 - 1s/epoch - 26ms/step
Epoch 18/100
51/51 - 1s - loss: 0.1133 - Accuracy: 0.9500 - val_loss: 0.1417 - val_Accuracy: 0.9426 - 1s/epoch - 26ms/step
Epoch 19/100
51/51 - 1s - loss: 0.1129 - Accuracy: 0.9519 - val_loss: 0.1372 - val_Accuracy: 0.9426 - 1s/epoch - 25ms/step
Epoch 20/100
51/51 - 1s - loss: 0.1117 - Accuracy: 0.9507 - val_loss: 0.1274 - val_Accuracy: 0.9426 - 1s/epoch - 26ms/step
Epoch 21/100
51/51 - 1s - loss: 0.1073 - Accuracy: 0.9572 - val_loss: 0.1299 - val_Accuracy: 0.9426 - 1s/epoch - 26ms/step
Epoch 22/100
51/51 - 1s - loss: 0.1132 - Accuracy: 0.9510 - val_loss: 0.1376 - val_Accuracy: 0.9438 - 1s/epoch - 26ms/step
Epoch 23/100
51/51 - 1s - loss: 0.1104 - Accuracy: 0.9532 - val_loss: 0.1203 - val_Accuracy: 0.9438 - 1s/epoch - 26ms/step
Epoch 24/100
51/51 - 1s - loss: 0.1062 - Accuracy: 0.9538 - val_loss: 0.1377 - val_Accuracy: 0.9413 - 1s/epoch - 26ms/step
Epoch 25/100
51/51 - 1s - loss: 0.1056 - Accuracy: 0.9538 - val_loss: 0.1269 - val_Accuracy: 0.9438 - 1s/epoch - 25ms/step
Epoch 26/100
51/51 - 1s - loss: 0.1077 - Accuracy: 0.9544 - val_loss: 0.1380 - val_Accuracy: 0.9426 - 1s/epoch - 25ms/step
Epoch 27/100
51/51 - 1s - loss: 0.1074 - Accuracy: 0.9532 - val_loss: 0.1152 - val_Accuracy: 0.9538 - 1s/epoch - 26ms/step
Epoch 28/100
51/51 - 1s - loss: 0.1049 - Accuracy: 0.9560 - val_loss: 0.1238 - val_Accuracy: 0.9488 - 1s/epoch - 27ms/step
Epoch 29/100
51/51 - 1s - loss: 0.1057 - Accuracy: 0.9525 - val_loss: 0.1159 - val_Accuracy: 0.9526 - 1s/epoch - 26ms/step
Epoch 30/100
51/51 - 1s - loss: 0.1084 - Accuracy: 0.9516 - val_loss: 0.1257 - val_Accuracy: 0.9488 - 1s/epoch - 26ms/step
Epoch 31/100
51/51 - 1s - loss: 0.1079 - Accuracy: 0.9538 - val_loss: 0.1125 - val_Accuracy: 0.9551 - 1s/epoch - 25ms/step
Epoch 32/100
51/51 - 1s - loss: 0.1067 - Accuracy: 0.9544 - val_loss: 0.1060 - val_Accuracy: 0.9538 - 1s/epoch - 27ms/step
Epoch 33/100
51/51 - 1s - loss: 0.1077 - Accuracy: 0.9569 - val_loss: 0.1097 - val_Accuracy: 0.9538 - 1s/epoch - 26ms/step
Epoch 34/100
51/51 - 1s - loss: 0.1036 - Accuracy: 0.9557 - val_loss: 0.1098 - val_Accuracy: 0.9538 - 1s/epoch - 26ms/step
Epoch 35/100
51/51 - 1s - loss: 0.1086 - Accuracy: 0.9572 - val_loss: 0.1164 - val_Accuracy: 0.9538 - 1s/epoch - 26ms/step
Epoch 36/100
51/51 - 1s - loss: 0.1038 - Accuracy: 0.9535 - val_loss: 0.1117 - val_Accuracy: 0.9576 - 1s/epoch - 26ms/step
Epoch 37/100
51/51 - 1s - loss: 0.1014 - Accuracy: 0.9591 - val_loss: 0.1049 - val_Accuracy: 0.9551 - 1s/epoch - 25ms/step
Epoch 38/100
51/51 - 1s - loss: 0.1007 - Accuracy: 0.9535 - val_loss: 0.1090 - val_Accuracy: 0.9563 - 1s/epoch - 26ms/step
Epoch 39/100
51/51 - 1s - loss: 0.1033 - Accuracy: 0.9579 - val_loss: 0.1126 - val_Accuracy: 0.9551 - 1s/epoch - 26ms/step
Epoch 40/100
51/51 - 1s - loss: 0.0996 - Accuracy: 0.9569 - val_loss: 0.1025 - val_Accuracy: 0.9588 - 1s/epoch - 28ms/step
Epoch 41/100
51/51 - 1s - loss: 0.1000 - Accuracy: 0.9579 - val_loss: 0.1009 - val_Accuracy: 0.9563 - 1s/epoch - 26ms/step
Epoch 42/100
51/51 - 1s - loss: 0.1064 - Accuracy: 0.9525 - val_loss: 0.0961 - val_Accuracy: 0.9625 - 1s/epoch - 26ms/step
Epoch 43/100
51/51 - 1s - loss: 0.1006 - Accuracy: 0.9554 - val_loss: 0.1014 - val_Accuracy: 0.9600 - 1s/epoch - 26ms/step
Epoch 44/100
51/51 - 1s - loss: 0.1000 - Accuracy: 0.9572 - val_loss: 0.1027 - val_Accuracy: 0.9576 - 1s/epoch - 27ms/step
Epoch 45/100
51/51 - 1s - loss: 0.1008 - Accuracy: 0.9541 - val_loss: 0.1034 - val_Accuracy: 0.9576 - 1s/epoch - 26ms/step
Epoch 46/100
51/51 - 1s - loss: 0.0976 - Accuracy: 0.9569 - val_loss: 0.1039 - val_Accuracy: 0.9551 - 1s/epoch - 26ms/step
Epoch 47/100
51/51 - 1s - loss: 0.1070 - Accuracy: 0.9522 - val_loss: 0.0952 - val_Accuracy: 0.9613 - 1s/epoch - 25ms/step
Epoch 48/100
51/51 - 1s - loss: 0.1002 - Accuracy: 0.9569 - val_loss: 0.0986 - val_Accuracy: 0.9588 - 1s/epoch - 26ms/step
Epoch 49/100
51/51 - 1s - loss: 0.1002 - Accuracy: 0.9575 - val_loss: 0.1024 - val_Accuracy: 0.9551 - 1s/epoch - 25ms/step
Epoch 50/100
51/51 - 1s - loss: 0.0980 - Accuracy: 0.9554 - val_loss: 0.0974 - val_Accuracy: 0.9613 - 1s/epoch - 27ms/step
Epoch 51/100
51/51 - 1s - loss: 0.0954 - Accuracy: 0.9575 - val_loss: 0.0977 - val_Accuracy: 0.9613 - 1s/epoch - 27ms/step
Epoch 52/100
51/51 - 1s - loss: 0.0941 - Accuracy: 0.9594 - val_loss: 0.0988 - val_Accuracy: 0.9613 - 1s/epoch - 27ms/step
Epoch 53/100
51/51 - 1s - loss: 0.1042 - Accuracy: 0.9554 - val_loss: 0.1000 - val_Accuracy: 0.9613 - 1s/epoch - 27ms/step
Epoch 54/100
51/51 - 1s - loss: 0.1009 - Accuracy: 0.9560 - val_loss: 0.0965 - val_Accuracy: 0.9613 - 1s/epoch - 26ms/step
Epoch 55/100
51/51 - 1s - loss: 0.0990 - Accuracy: 0.9575 - val_loss: 0.0967 - val_Accuracy: 0.9613 - 1s/epoch - 26ms/step
Epoch 56/100
51/51 - 1s - loss: 0.0967 - Accuracy: 0.9585 - val_loss: 0.0944 - val_Accuracy: 0.9613 - 1s/epoch - 26ms/step
Epoch 57/100
51/51 - 1s - loss: 0.0956 - Accuracy: 0.9579 - val_loss: 0.0929 - val_Accuracy: 0.9600 - 1s/epoch - 27ms/step
Epoch 58/100
51/51 - 1s - loss: 0.0973 - Accuracy: 0.9582 - val_loss: 0.0921 - val_Accuracy: 0.9613 - 1s/epoch - 26ms/step
Epoch 59/100
51/51 - 1s - loss: 0.0996 - Accuracy: 0.9566 - val_loss: 0.0937 - val_Accuracy: 0.9613 - 1s/epoch - 25ms/step
Epoch 60/100
51/51 - 1s - loss: 0.0958 - Accuracy: 0.9579 - val_loss: 0.0958 - val_Accuracy: 0.9600 - 1s/epoch - 25ms/step
Epoch 61/100
51/51 - 1s - loss: 0.1043 - Accuracy: 0.9557 - val_loss: 0.0912 - val_Accuracy: 0.9613 - 1s/epoch - 27ms/step
Epoch 62/100
51/51 - 1s - loss: 0.0952 - Accuracy: 0.9597 - val_loss: 0.0928 - val_Accuracy: 0.9600 - 1s/epoch - 25ms/step
Epoch 63/100
51/51 - 1s - loss: 0.0964 - Accuracy: 0.9569 - val_loss: 0.0932 - val_Accuracy: 0.9600 - 1s/epoch - 26ms/step
Epoch 64/100
51/51 - 1s - loss: 0.0920 - Accuracy: 0.9607 - val_loss: 0.0921 - val_Accuracy: 0.9600 - 1s/epoch - 26ms/step
Epoch 65/100
51/51 - 1s - loss: 0.0947 - Accuracy: 0.9566 - val_loss: 0.0923 - val_Accuracy: 0.9613 - 1s/epoch - 25ms/step
Epoch 66/100
51/51 - 1s - loss: 0.0935 - Accuracy: 0.9588 - val_loss: 0.0924 - val_Accuracy: 0.9613 - 1s/epoch - 26ms/step
Epoch 67/100
51/51 - 1s - loss: 0.0950 - Accuracy: 0.9597 - val_loss: 0.0920 - val_Accuracy: 0.9600 - 1s/epoch - 26ms/step
Epoch 68/100
51/51 - 1s - loss: 0.0933 - Accuracy: 0.9554 - val_loss: 0.0922 - val_Accuracy: 0.9600 - 1s/epoch - 26ms/step
Epoch 69/100
51/51 - 1s - loss: 0.0940 - Accuracy: 0.9554 - val_loss: 0.0944 - val_Accuracy: 0.9600 - 1s/epoch - 25ms/step
Epoch 70/100
51/51 - 1s - loss: 0.0981 - Accuracy: 0.9566 - val_loss: 0.0888 - val_Accuracy: 0.9613 - 1s/epoch - 26ms/step
Epoch 71/100
51/51 - 1s - loss: 0.0918 - Accuracy: 0.9607 - val_loss: 0.0911 - val_Accuracy: 0.9613 - 1s/epoch - 26ms/step
Epoch 72/100
51/51 - 1s - loss: 0.0935 - Accuracy: 0.9563 - val_loss: 0.0920 - val_Accuracy: 0.9600 - 1s/epoch - 26ms/step
Epoch 73/100
51/51 - 1s - loss: 0.0966 - Accuracy: 0.9560 - val_loss: 0.0923 - val_Accuracy: 0.9588 - 1s/epoch - 26ms/step
Epoch 74/100
51/51 - 1s - loss: 0.0930 - Accuracy: 0.9575 - val_loss: 0.0905 - val_Accuracy: 0.9600 - 1s/epoch - 26ms/step
Epoch 75/100
51/51 - 1s - loss: 0.0930 - Accuracy: 0.9600 - val_loss: 0.0906 - val_Accuracy: 0.9600 - 1s/epoch - 26ms/step
Epoch 76/100
51/51 - 1s - loss: 0.0968 - Accuracy: 0.9566 - val_loss: 0.0961 - val_Accuracy: 0.9613 - 1s/epoch - 25ms/step
Epoch 77/100
51/51 - 1s - loss: 0.0888 - Accuracy: 0.9591 - val_loss: 0.0932 - val_Accuracy: 0.9625 - 1s/epoch - 25ms/step
Epoch 78/100
51/51 - 1s - loss: 0.0929 - Accuracy: 0.9597 - val_loss: 0.0900 - val_Accuracy: 0.9613 - 1s/epoch - 26ms/step
Epoch 79/100
51/51 - 1s - loss: 0.0916 - Accuracy: 0.9607 - val_loss: 0.0899 - val_Accuracy: 0.9576 - 1s/epoch - 25ms/step
Epoch 80/100
51/51 - 1s - loss: 0.0920 - Accuracy: 0.9613 - val_loss: 0.0920 - val_Accuracy: 0.9588 - 1s/epoch - 25ms/step
Epoch 81/100
51/51 - 1s - loss: 0.0922 - Accuracy: 0.9588 - val_loss: 0.0900 - val_Accuracy: 0.9613 - 1s/epoch - 26ms/step
Epoch 82/100
51/51 - 1s - loss: 0.0972 - Accuracy: 0.9560 - val_loss: 0.0888 - val_Accuracy: 0.9613 - 1s/epoch - 25ms/step
----
train shape: (3203, 600, 1)
score on val: 0.961298406124115
score on train: 0.9644083380699158
----
training time (s): 113.8620
Plot training and evaluate the model#
We can plot the training and validation accuracy and loss to visualize the training progress.
acc = history.history['Accuracy']
val_acc = history.history['val_Accuracy']
epochs = range(1, len(acc) + 1)
fig, axs = plt.subplots(1, 2, figsize=(8, 4))
axs[0].plot(epochs, acc, c='#12B5CB', label='Training')
axs[0].plot(epochs, val_acc, c='#425066', label='Validation')
axs[0].set_title('Training and validation accuracy\n')
axs[0].set_xlabel('Epochs')
axs[0].set_ylabel('Accuracy')
axs[0].legend()
loss = history.history['loss']
val_loss = history.history['val_loss']
axs[1].plot(epochs, loss, c='#12B5CB', label='Training')
axs[1].plot(epochs, val_loss, c='#425066', label='Validation')
axs[1].set_title('Training and validation loss\n')
axs[1].set_xlabel('Epochs')
axs[1].set_ylabel('Loss')
axs[1].legend()
plt.tight_layout()
plt.show()
best_epoch = val_acc.index(max(val_acc)) + 1
print(f'Best epoch: {best_epoch} (accuracy={max(val_acc):.4f})')
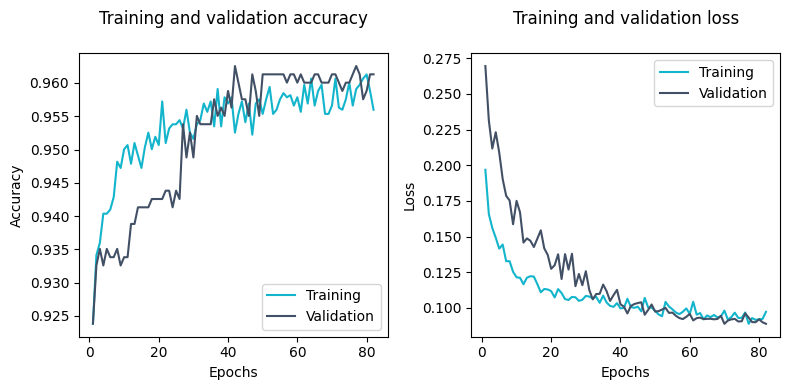
Best epoch: 42 (accuracy=0.9625)
We can plot the ROC curve and the confusion matrix to further evaluate our trained model.
y_preds = new_model.predict(x_test, verbose=0).ravel()
fpr, tpr, thresholds = roc_curve(y_test, y_preds)
fig, axs = plt.subplots(1, 2, figsize=(8, 4))
axs[0].plot([0, 1], [0, 1], 'y--')
axs[0].plot(fpr, tpr, marker='.')
axs[0].set_xlabel('False positive rate')
axs[0].set_ylabel('True positive rate')
axs[0].set_title('ROC curve\n')
optimal_threshold = thresholds[np.argmax(tpr - fpr)]
y_pred2 = (new_model.predict(x_test, verbose=0) >= optimal_threshold).astype(int)
cm = confusion_matrix(y_test, y_pred2)
disp = ConfusionMatrixDisplay(confusion_matrix=cm)
disp.plot(cmap='Blues', ax=axs[1])
axs[1].set_title('Confusion matrix\n')
plt.tight_layout()
plt.show()
print(f'Area under curve, AUC = {auc(fpr, tpr)}')
print(f'Optimal threshold value = {optimal_threshold}')
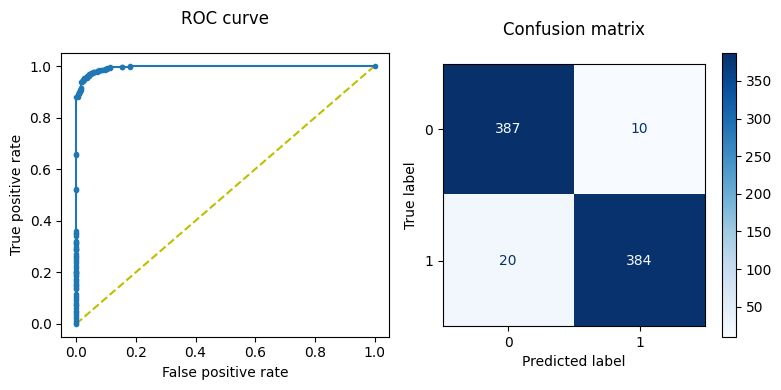
Area under curve, AUC = 0.9953612489712447
Optimal threshold value = 0.648914635181427
Save trained model and training settings#
Finally, we save the trained model to an .h5 file and the training settings to a text file.
new_model.save('../results/lstm_transfer.h5')
with open('../results/tl_training_settings.txt', 'w') as text_file:
text_file.write('\n'.join(f'{i}: {settings[i]}' for i in settings))
/opt/miniconda3/envs/miniml/lib/python3.9/site-packages/keras/src/engine/training.py:3103: UserWarning: You are saving your model as an HDF5 file via `model.save()`. This file format is considered legacy. We recommend using instead the native Keras format, e.g. `model.save('my_model.keras')`.
saving_api.save_model(
