Loading data#
To import data into miniML, we first need to import miniML’s class MiniTrace.
import sys
sys.path.append('../../core/')
from miniML import MiniTrace
miniML operates on NumPy ndarrays and we can use any 1-dimensional array for event detection with miniML. In addition, miniML includes functions for loading data from various file formats that are commonly used in electrophysiology. These include HEKA PatchMaster (.dat), Axon Binary Files (.abf), and .h5 files. Data in other formats can be imported into Python as a NumPy array.
Loading data from .h5 files#
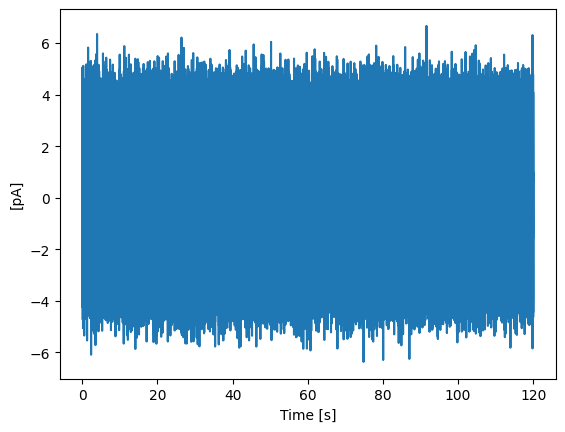
Here, we will load data from a .h5 file. The loading is done using the from_h5_file() method of the MiniTrace class. The required arguments are filename, tracename, scaling, sampling, and unit.
filename = '../../example_data/gc_mini_trace.h5'
scaling = 1e12
unit = 'pA'
# get from h5 file
h5_trace = MiniTrace.from_h5_file(filename=filename,
tracename='mini_data',
scaling=scaling,
sampling=2e-5,
unit=unit)
h5_trace.plot_trace()
Loading data from .abf files#
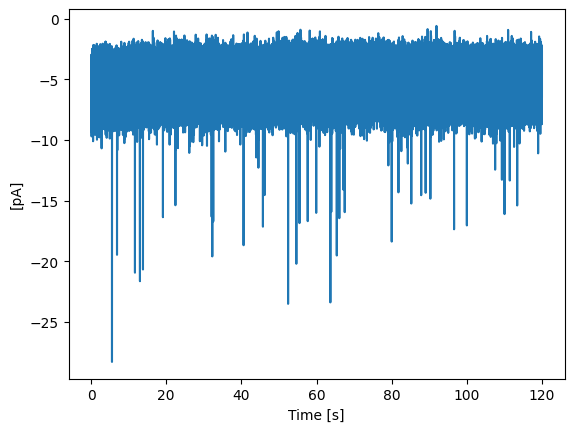
Data can also be loaded from Axon binary files (.abf). The .abf import method from_axon_file() makes use of the pyAbf library.
filename = '../../example_data/gc_mini_trace.abf'
scaling = 1
unit = 'pA'
# get from ABF file
abf_trace = MiniTrace.from_axon_file(filename=filename,
channel=0,
scaling=scaling,
unit=unit)
abf_trace.plot_trace()
Loading data from .dat files#
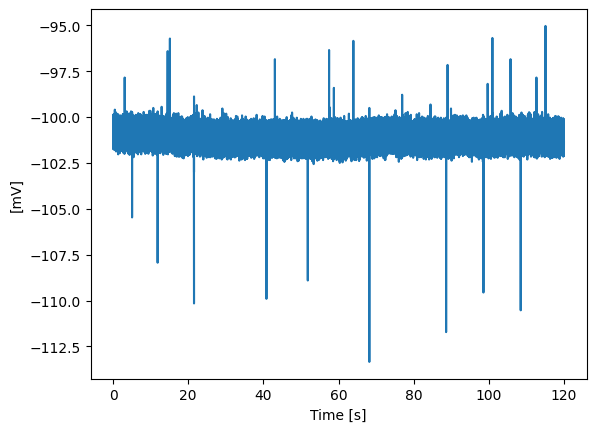
To load data from HEKA PatchMaster (.dat) files, use the from_heka_file() method. You need to specify the PatchMaster PGF name of the recording you want to load using the rectype argument. Optional arguments are group, scaling and unit.
See also
Please refer to the from_heka_file() documentation for more information.
filename = '../../example_data/B_2020-05-14_007.dat'
rectype = 'conti CC'
scaling = 1e3
unit = 'mV'
PM_trace = MiniTrace.from_heka_file(filename=filename,
rectype=rectype,
group=1,
scaling=scaling,
unit=unit)
PM_trace.plot_trace()
Loading data from other sources#
To analyse data from any other file format, you need to import the recording into Python as a NumPy array. NumPy can read various file formats such as .csv, .txt, etc.
Any 1d array can then be used to initiate a miniML MiniTrace object. Here, we create a NumPy array containing white noise. When creating a MiniTrace object, the required arguments are data, sampling_interval, y_unit, and filename.
import numpy as np
example_data = np.random.normal(0, 1.25, 6_000_000)
np_trace = MiniTrace(data=example_data,
sampling_interval=2e-5,
y_unit='pA',
filename=None)
np_trace.plot_trace()